White PaperArticle
The LumiraDx SARS-CoV-2 Antigen Test aligns with detection of infectious samples from immunocompetent individuals with none to mild and moderate symptoms
Background:
Since COVID-19 was declared as a pandemic by the World Health Organization (WHO) in March 2020, over one hundred million cases and almost 3 million deaths have occurred worldwide (1). The etiologic agent (SARS-CoV-2) spreads readily from person-to-person mainly by respiratory transmission (2). Transmission requires a minimal dose of replication competent virus delivered to a susceptible host. Therefore, individuals actively shedding live virus are suspected to be infectious. To mitigate spread of the virus, isolation of individuals with confirmed infections (and their close contacts), as well as the restriction of overall social movement, is often mandated. These measures have significant consequences for societies and therefore, it is of paramount importance to understand the relationship between viral detection and the ability to transmit disease. This paper presents a summary of available literature on that topic.
Symptom based testing/ Days since symptom onset (DSSO)
To determine the optimal duration of isolation for individuals with confirmed SARS-CoV-2 infections, one needs to know how long people are spreading viable virus, i.e. infectious. Many studies have been performed to address this question and they demonstrate that people with mild-moderate symptoms are unlikely to be infective longer than 10 DSSO (4-8). Based on those findings, the Centres of Disease Control and Prevention (CDC), as well as the European Centre for Disease Prevention and Control (ECDC) have stated that isolation of COVID-19 patients with mild to moderate symptoms can be discontinued after 10 DSSO, if they also show resolution of fewer and improvement in their overall symptoms for at least 1 or 3 days, respectively (9,10).
Viral cell culture
The current understanding of SARS-CoV-2 transmission is mainly based on substantial evidence derived from contact and contamination tracing, as well as viral cell culture-based studies (7, 8, 2). Viral culture is commonly used as surrogate indicator for the infective potential of the virus (6, 8, 12). Specimen from SARS-CoV-2 RNA positive individuals is transferred to viral cell culture in order to investigate if replication-competent virus can be detected. In addition, samples collected from individuals with mild-moderate symptoms at DSSO>10 are only cell culture positive in a few cases, compared to the high positivity rate when specimens are collected earlier in the disease course. Therefore, it has been concluded that individuals who are > 10 DSSO are highly unlikely to be infective (4-8). Furthermore, cell culture positivity increases with higher viral loads that are more common earlier in the infection (5, 12, 13). Antigen testing methods align more commonly with viral culture than reverse transcriptase Polymerase Chain Reaction (rt-PCR) (14, 20). Both methods and the link between viral culture and infectivity are described below.
Reverse transcriptase Polymerase Chain Reaction (rt-PCR) for detection of SARS-CoV-2 infection
One of the methods used for detection of SARS-CoV-2 infected individuals is rt-PCR. This method detects the amount of viral RNA in specimens usually collected from nasal samples, anterior nares or nasopharyngeal swabs. This method was the first tool utilized for the reliable detection of SARS-CoV-2 (3) and is still considered the gold standard for COVID-19 diagnosis, often serving as the reference method for other detection systems. Results are often presented as cycle threshold (Ct) values. The higher the Ct value, the lower the amount of RNA copies in the analysed sample. Ct values typically range from 10–40. However, detection of RNA alone does not necessarily translate to the presence of virus with infective potential, and samples with a Ct>33 have been demonstrated to be predominantly cell culture negative for SARS-CoV-2 (suggesting, that only samples with CT<33-34 should be considered as potentially infectious (13)).
CT values are semiquantitative and vary widely between rt-PCR systems making comparisons difficult (22, 23). In order to improve this, some studies have converted their rt-PCR results as viral copies/ml, usually called viral load (14, 24). Viral load and viral RNA concentration are proportional and samples with higher values are more likely to be SARS-CoV-2 cell culture positive. Based on the current literature, samples with more than 6 log10 viral RNA copies/ml are likely to correlate with SARS-CoV-2 cell culture positivity (CCP) (5, 12, 14, 16).
Rt-PCR methods are very sensitive and can detect even small amounts of RNA. However, this is not necessarily an optimal method for prediction of infectivity of an individual, since it detects all RNA, including remnant RNA which may not always be representative of live, replicating virus (5, 15, 25, 26). Remnant RNA can persist in some people for up to 109 days (26). As such, positive rt-PCR results that may not be representative of the ability to transmit disease and could confound by overestimating the infective window and/or delay an individuals’ return to work/ school. Furthermore, this method requires skilled laboratory technologists, cannot be performed at the point of care, is labour and time intensive, and often expensive.
Detection of SARS-CoV-2 antigen
Another method used to detect SARS-CoV-2 infection is detection of the SARS-CoV-2 antigen in specimens collected by nasal or nasopharyngeal swabbing. In these methods, the SARS-CoV-2 nucleocapsid protein is detected and bound by specific antibodies from which a threshold level of antigen that correlates to positive infection has been previously determined. Most of the available antigen test kits are based on the lateral flow principle. These tests are not as sensitive as rt-PCR but can detect antigen in highly infective individuals and some can detect the majority of individuals up to Ct 25-30 (16- 19). Most of those tests are read by visual inspection of the test line, present a result between 15-30 minutes, and can be performed outside of a conventional clinical diagnostic laboratory setting. However, infectious individuals with higher Cts, up to the infectious window of 30≤Ct<33, are mostly missed with these tests (17).
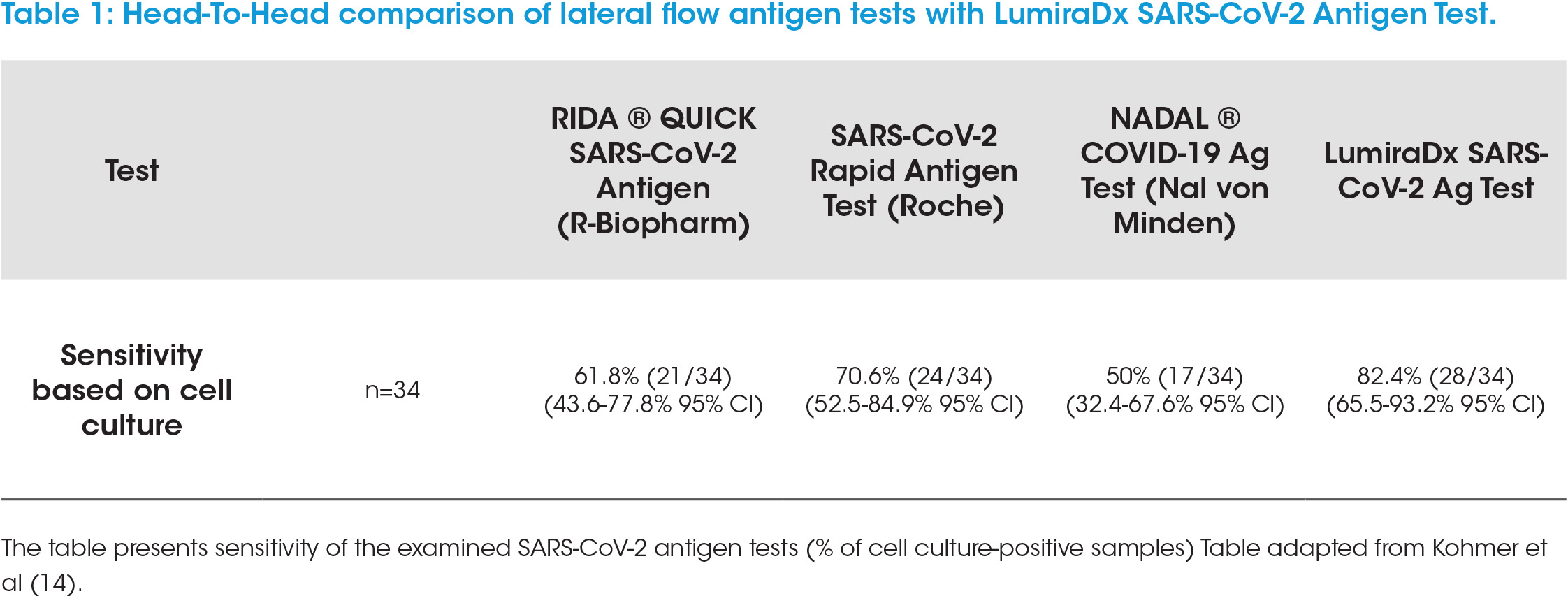
A newer technology, based on rapid microfluidic immunofluorescence, is the LumiraDx SARS-CoV-2 Antigen Test. This convenient point of care test system allows rapid, qualitative detection of nucleocapsid protein antigen within 12 minutes of sample application. The test has a very low limit of detection (LOD) (32 TCID50/ml) and can determine positive samples up to 12 DSSO and Ct<33 (Roche Cobas) (11). Independent prospective studies have confirmed the test sensitivity (21). Head-to-head comparisons with lateral flow tests have demonstrated that the LumiraDx SARS-CoV-2 Antigen Test detects clearly more SARS-CoV-2 cell culture positive samples (Table 1, adapted from Kohmer et al. 14) and demonstrates a superior sensitivity by lower LOD (24). Taken together, this shows that the LumiraDx SARS-CoV-2 antigen test detects samples which are likely to be in the infectious window (≤12 DSSO and samples with Ct<33) and is more sensitive, by means of a lower LOD, than lateral flow tests.
Relevance
It is of major importance to detect infectious individuals in a sensitive and reliable manner to prevent the spread of infection, manage infectious patients appropriately in health systems and overall to enable normal life to resume. Rt-PCR is considered the gold standard for detection of SARS-CoV-2 infection; however, it detects RNA irrespective of the infective potential of the individual. SARS-CoV-2 antigen detection is representative of the presence of viable virus, but not all antigen tests are sensitive enough to determine the wider window of infectivity. Lack of test sensitivity can risk some infectious individuals being missed especially by usage of lateral flow (LF) tests with sub-optimal LODs. In particular, the very high sensitivity of the LumiraDx SARS-CoV-2 Antigen Test enables it to be used in situations where infection control is critical. For example, it is an outstanding tool for supporting admission control in hospitals, GPs or Care Homes, management as well as safe discharge of patients from hospital, work-place testing such as for emergency services or in remote locations such as Oil and Gas rigs (27-31).
Summary
The LumiraDx SARS-CoV-2 Antigen Test has superior analytical sensitivity in direct comparison to lateral flow tests (32) and detects samples which are more likely to be in the infectious window (≤12 DSSO and Ct<33), while avoiding the detection of non-viable virus, as seen with rt-PCR. Therefore, it meets a so far unmet need and reduces the gap between individuals who are missed by LF tests or overestimated by rt-PCR. Additionally, the LumiraDx SARS-CoV-2 Antigen Test can be used at the point of care giving a rapid result in 12 minutes. In summary, the LumiraDx SARS-CoV-2 Antigen Test represents an important tool to support appropriate patient management and for detection of infectious individuals.
References:
2. www.ncbi.nlm.nih.gov/pmc/articles/PMC7505025/
3. www.ncbi.nlm.nih.gov/pmc/articles/PMC6988269/
4. www.ncbi.nlm.nih.gov/pmc/articles/PMC7427302/
5. www.nature.com/articles/s41586-020-2196-x
6.www.ncbi.nlm.nih.gov/pmc/articles/PMC7665383/pdf/ciaa1579.pdf
7. www.ncbi.nlm.nih.gov/pmc/articles/PMC7547320/pdf/main.pdf
8. www.hiqa.ie/reports-and-publications/health-technology-assessment/duration-infectiousness-sars-cov-2
9. www.ecdc.europa.eu/sites/default/files/documents/Guidance-for-discharge-and-ending-of-isolation-ofpeople-with-COVID-19.pdf
10. www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html
11. https://link.springer.com/article/10.1007/s40121-021-00413-x
12. www.nature.com/articles/s41467-020-20568-4
13. www.ncbi.nlm.nih.gov/pmc/articles/PMC7185831/
14. www.ncbi.nlm.nih.gov/pmc/articles/PMC7830733/
15. www.thelancet.com/action/showPdf?pii=S2352-3964%2820%2930336-4
16. www.hug.ch/sites/interhug/files/structures/laboratoire_de_virologie/documents/Centre_maladies_virales_ infectieuses/ofsp_rdt_report_gcevd_27.10.2020.pdf
17. www.synlab.com/human/news/news-article/medical-data-reveal-that-almost-40-of-sars-cov-2-carriers-canbe-missed-by-rapid-antigen-tests-91
18. https://clinical.r-biopharm.com/wp-content/uploads/sites/3/2020/09/n6803_ridaquick-sars-cov-2_antigen_2020-09-10_de.pdf
19. www.sciencedirect.com/science/article/pii/S1386653220304558
20. https://academic.oup.com/cid/advance-article/doi/10.1093/cid/ciaa1706/6105729
21. https://skup.org/
22. https://www.sciencemag.org/news/2020/09/one-number-could-help-reveal-how-infectious-covid-19-patient-should-test-results
23. https://research.uiowa.edu/sites/research.uiowa.edu/files/ct_value_cap_summary.pdf
24. www.medrxiv.org/content/10.1101/2021.03.02.21252430v1
25. https://pubmed.ncbi.nlm.nih.gov/33180094/
26. www.thelancet.com/action/showPdf?pii=S2352-3964%2821%2900023-2
27. www.medrxiv.org/content/10.1101/2021.04.22.21255948v2.full
28. www.lumiradx.com/uk-en/case-studies/lumiradx-sars-cov-2-antigen-test-supports-established-testingconcepts-with-rapid-and-accurate-results
29. www.lumiradx.com/uk-en/case-studies/lumiradx-sars-cov-2-antigen-test-supports-operational-readiness-offire-brigades-in-lower-austria
30. www.lumiradx.com/uk-en/case-studies/use-of-the-lumiradx-sars-cov-2-antigen-test-in-pediatric-practice
31. www.energyvoice.com/coronavirus/265546/aberdeen-clinic-tests-coronavirus/
32. www.lumiradx.com/assets/pdfs/covid-19-antigen-test/sars-cov-2-ag-test-superior-analytical-sensitivity.pdf?v=1